3. Alternative polyadenylation
shift.RmdAPA sites can be detected using the PAT-Seq protocol. This protocol produces 3’-end focussed reads. We examine GSE83162. This is a time-series experiment in which two strains of yeast are released into synchronized cell cycling and observed through two cycles. Yeast are treated with \(\alpha\)-factor, which causes them to stop dividing in antici… pation of a chance to mate. When the \(\alpha\)-factor is washed away, they resume cycling.
Load files
library(tidyverse)
library(reshape2)
library(limma)
library(topconfects)
library(weitrix)
peaks <- system.file("GSE83162", "peaks.csv.gz", package="weitrix") %>%
read_csv()
counts <- system.file("GSE83162", "peak_count.csv.gz", package="weitrix") %>%
read_csv() %>%
column_to_rownames("name") %>%
as.matrix()
genes <- system.file("GSE83162", "genes.csv.gz", package="weitrix") %>%
read_csv() %>%
column_to_rownames("name")
samples <- data.frame(sample=I(colnames(counts))) %>%
extract(sample, c("strain","time"), c("(.+)-(.+)"), remove=FALSE) %>%
mutate(
strain=factor(strain,unique(strain)),
time=factor(time,unique(time)))
rownames(samples) <- samples$sample
samples## sample strain time
## WT-tpre WT-tpre WT tpre
## WT-t0m WT-t0m WT t0m
## WT-t15m WT-t15m WT t15m
## WT-t30m WT-t30m WT t30m
## WT-t45m WT-t45m WT t45m
## WT-t60m WT-t60m WT t60m
## WT-t75m WT-t75m WT t75m
## WT-t90m WT-t90m WT t90m
## WT-t105m WT-t105m WT t105m
## WT-t120m WT-t120m WT t120m
## DeltaSet1-tpre DeltaSet1-tpre DeltaSet1 tpre
## DeltaSet1-t0m DeltaSet1-t0m DeltaSet1 t0m
## DeltaSet1-t15m DeltaSet1-t15m DeltaSet1 t15m
## DeltaSet1-t30m DeltaSet1-t30m DeltaSet1 t30m
## DeltaSet1-t45m DeltaSet1-t45m DeltaSet1 t45m
## DeltaSet1-t60m DeltaSet1-t60m DeltaSet1 t60m
## DeltaSet1-t75m DeltaSet1-t75m DeltaSet1 t75m
## DeltaSet1-t90m DeltaSet1-t90m DeltaSet1 t90m
## DeltaSet1-t105m DeltaSet1-t105m DeltaSet1 t105m
## DeltaSet1-t120m DeltaSet1-t120m DeltaSet1 t120mgroups <- select(peaks, group=gene_name, name=name)
# Note the order of this data frame is important
wei <- counts_shift(counts, groups)## Calculating shifts in 1 blocksExploratory analysis
We can look for components of variation.
comp_seq <- weitrix_components_seq(wei, p=10, design=~0)components_seq_screeplot(comp_seq)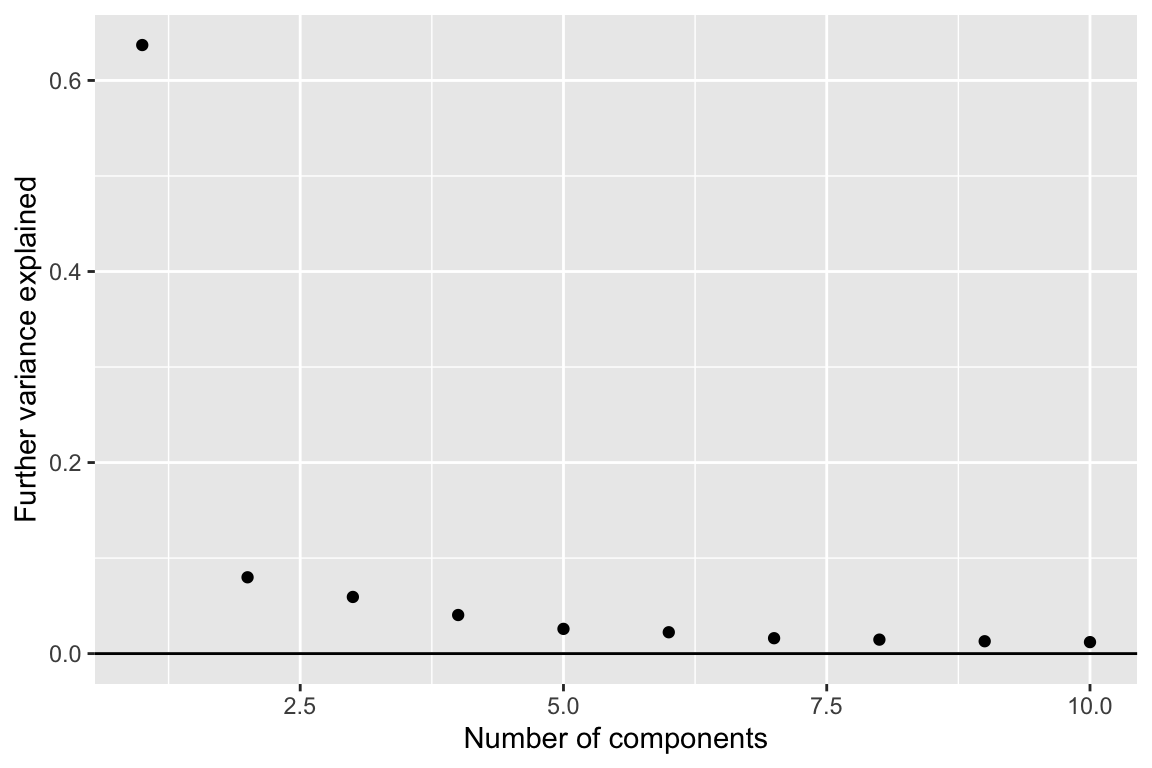
Pushing a point somewhat, we examine four components.
comp <- comp_seq[[4]]
comp$col %>% melt(varnames=c("sample","component")) %>%
left_join(samples,by="sample") %>%
ggplot(aes(x=time, y=value, color=strain, group=strain)) +
geom_hline(yintercept=0) +
geom_line() +
geom_point(alpha=0.75, size=3) +
facet_grid(component ~ .) +
labs(title="Sample scores for each component", y="Sample score", x="Time", color="Strain")## Warning: Column `sample` joining factor and character vector, coercing into character vector## Warning: Column `sample` has different attributes on LHS and RHS of join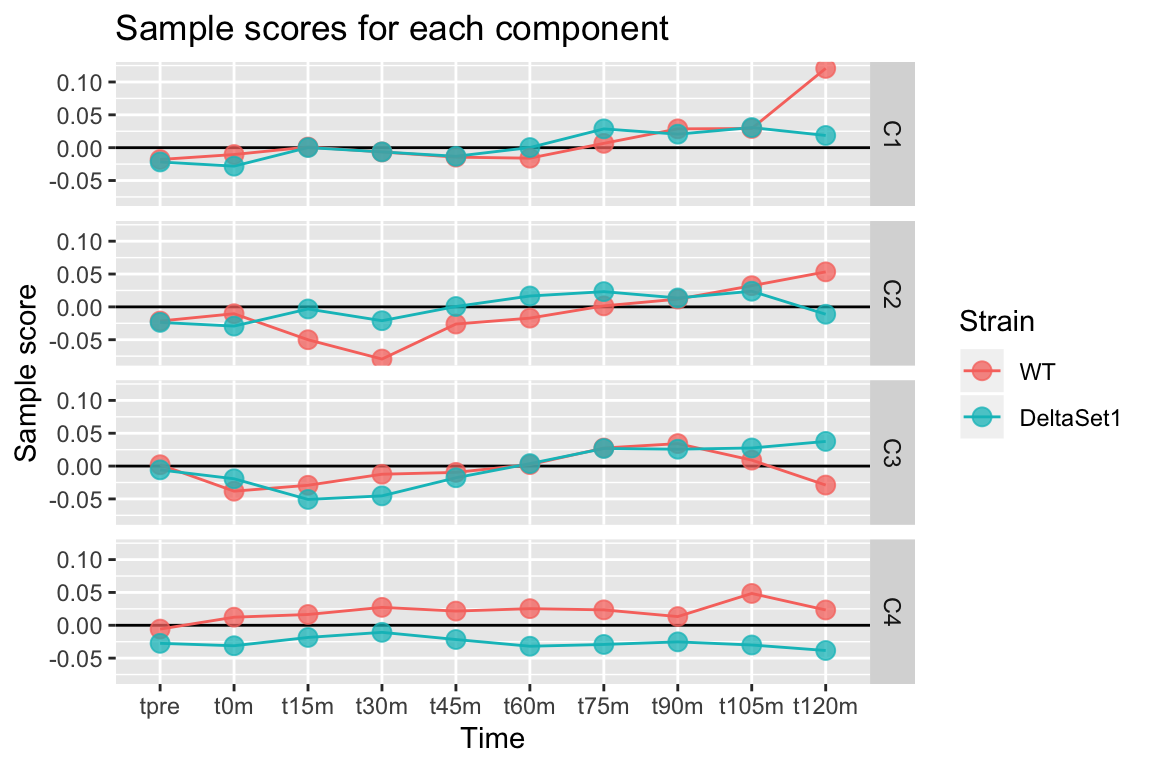
A weitrix created with counts_shift has a built-in default trend formula, so we don’t need to give a formula explicitly to weitrix_calibrate_trend.
cal_comp <- weitrix_calibrate_trend(wei, comp)
metadata(cal_comp)$weitrix$trend_fit##
## Call:
## lm(formula = log(dispersion_before) ~ log(per_read_var) + splines::ns(log(total_reads),
## 3), data = data)
##
## Coefficients:
## (Intercept) log(per_read_var) splines::ns(log(total_reads), 3)1 splines::ns(log(total_reads), 3)2 splines::ns(log(total_reads), 3)3
## 0.661 1.292 1.258 3.431 5.302fit_comp <- cal_comp %>%
weitrix_elist() %>%
lmFit(comp$col)Treat these results with caution. Confindence bounds take into account uncertainty in the loadings but not in the scores! What follows is best regarded as exploratory rather than a final result.
Gene loadings for C1
limma_confects(fit_comp, "C1", full=TRUE, fdr=0.05)## $table
## rank index confect effect se df fdr_zero AveExpr name per_read_var total_reads symbol biotype product dispersion_before dispersion_trend dispersion_after
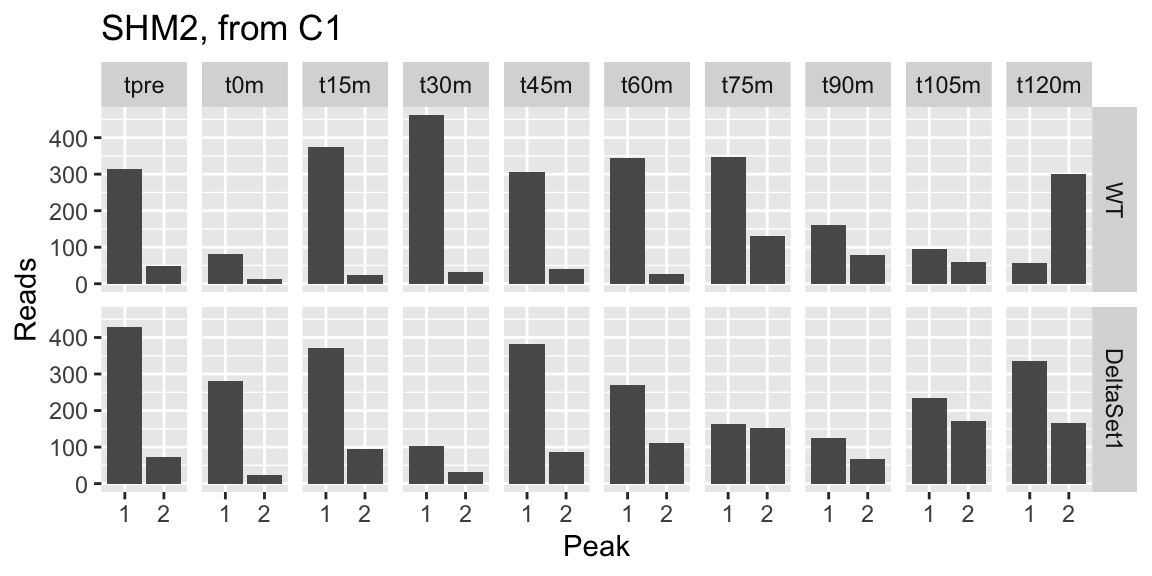
## 1 1134 2.622 4.094607 0.3234224 37.13126 1.456417e-12 0.0081521309 YLR058C 0.18681547 6958 SHM2 protein_coding Cytosolic serine hydroxymethyltransferase; converts serine to glycine plus 5,10 methylenetetrahydrofolate; major isoform involved in generating precursors for purine, pyrimidine, amino acid, and lipid biosynthesis [Source:SGD;Acc:S000004048] 0.4715635 0.5378079 0.8768252
## 2 1767 -2.283 -3.305108 0.2363955 37.13126 1.022366e-13 0.0034781506 YPR080W 0.20630257 72240 TEF1 protein_coding Translational elongation factor EF-1 alpha; also encoded by TEF2; functions in the binding reaction of aminoacyl-tRNA (AA-tRNA) to ribosomes; may also have a role in tRNA re-export from the nucleus; TEF1 has a paralog, TEF2, that arose from the whole genome duplication [Source:SGD;Acc:S000006284] 2.8807828 2.0867493 1.3805122
## 3 1033 -2.010 -2.761523 0.1792251 37.13126 1.459166e-14 -0.0132153436 YKL096W-A 0.08003464 374176 CWP2 protein_coding Covalently linked cell wall mannoprotein; major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored [Source:SGD;Acc:S000001956] 12.4806180 1.7432498 7.1593974
## 4 169 1.967 3.270320 0.3184946 37.13126 4.806902e-10 0.0091149690 YBR127C 0.19375593 21804 VMA2 protein_coding Subunit B of V1 peripheral membrane domain of vacuolar H+-ATPase; an electrogenic proton pump found throughout the endomembrane system; contains nucleotide binding sites; also detected in the cytoplasm; protein abundance increases in response to DNA replication stress [Source:SGD;Acc:S000000331] 1.7285216 0.9814567 1.7611797
## 5 114 1.847 2.588946 0.1847195 37.13126 1.022366e-13 0.0046972242 YBL072C 0.12048014 47370 RPS8A protein_coding Protein component of the small (40S) ribosomal subunit; homologous to mammalian ribosomal protein S8, no bacterial homolog; RPS8A has a paralog, RPS8B, that arose from the whole genome duplication [Source:SGD;Acc:S000000168] 0.7234498 0.8136384 0.8891540
## 6 1261 1.772 2.630669 0.2169606 37.13126 4.576818e-12 0.0098433471 YML026C 0.11684482 42444 RPS18B protein_coding Protein component of the small (40S) ribosomal subunit; homologous to mammalian ribosomal protein S18 and bacterial S13; RPS18B has a paralog, RPS18A, that arose from the whole genome duplication; protein abundance increases in response to DNA replication stress [Source:SGD;Acc:S000004488] 1.2292025 0.7346213 1.6732464
## 7 442 1.385 3.766530 0.6118541 37.13126 2.750072e-05 0.0005048926 YDR342C 0.16037644 2866 HXT7 protein_coding High-affinity glucose transporter; member of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels; HXT7 has a paralog, HXT4, that arose from the whole genome duplication [Source:SGD;Acc:S000002750] 0.8407210 0.3078081 2.7313154
## 8 502 1.385 1.979957 0.1542252 37.13126 1.143652e-12 0.0089272010 YDR524C-B 0.12463621 164069 <NA> protein_coding Putative protein of unknown function; YDR524C-B has a paralog, YCL048W-A, that arose from the whole genome duplication [Source:SGD;Acc:S000028739] 1.8409114 1.8042278 1.0203321
## 9 1060 -1.346 -2.294213 0.2483615 37.13126 6.646478e-09 -0.0212733375 YKL180W 0.17678312 57320 RPL17A protein_coding Ribosomal 60S subunit protein L17A; copurifies with the Dam1 complex (aka DASH complex); homologous to mammalian ribosomal protein L17 and bacterial L22; RPL17A has a paralog, RPL17B, that arose from the whole genome duplication; protein abundance increases in response to DNA replication stress [Source:SGD;Acc:S000001663] 1.3279041 1.4911642 0.8905150
## 10 210 1.250 2.282676 0.2730356 37.13126 6.425037e-08 0.0138000877 YBR249C 0.16608558 15246 ARO4 protein_coding 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase; catalyzes the first step in aromatic amino acid biosynthesis and is feedback-inhibited by tyrosine or high concentrations of phenylalanine or tryptophan; relative distribution to the nucleus increases upon DNA replication stress [Source:SGD;Acc:S000000453] 0.3423060 0.6693189 0.5114243
## ...
## 109 of 1805 non-zero log2 fold change at FDR 0.05
## Prior df 21.1Gene loadings for C2
limma_confects(fit_comp, "C2", full=TRUE, fdr=0.05)## $table
## rank index confect effect se df fdr_zero AveExpr name per_read_var total_reads symbol biotype product dispersion_before dispersion_trend dispersion_after
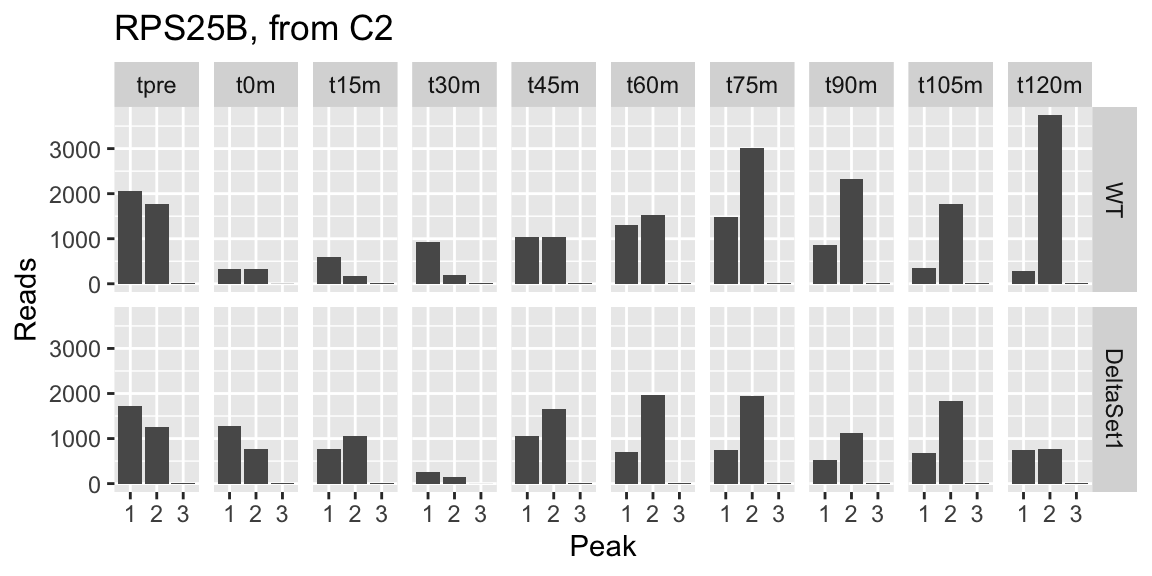
## 1 1218 4.703 6.337109 0.3649066 37.13126 1.722925e-16 -0.048806064 YLR333C 0.2382249 46413 RPS25B protein_coding Protein component of the small (40S) ribosomal subunit; homologous to mammalian ribosomal protein S25, no bacterial homolog; RPS25B has a paralog, RPS25A, that arose from the whole genome duplication [Source:SGD;Acc:S000004325] 1.6666532 1.9407819 0.8587535
## 2 1111 4.703 6.283793 0.3654725 37.13126 1.722925e-16 -0.058877133 YLL024C 0.1959787 133748 SSA2 protein_coding ATP-binding protein; involved in protein folding and vacuolar import of proteins; member of heat shock protein 70 (HSP70) family; associated with the chaperonin-containing T-complex; present in the cytoplasm, vacuolar membrane and cell wall; 98% identical with paralog Ssa1p, but subtle differences between the two proteins provide functional specificity with respect to propagation of yeast [URE3] prions and vacuolar-mediated degradations of gluconeogenesis enzymes [Source:SGD;Acc:S000003947] 5.1568575 2.8464338 1.8116906
## 3 1145 4.147 4.861895 0.1705474 37.13126 1.374939e-23 -0.041241296 YLR110C 0.2070177 712909 CCW12 protein_coding Cell wall mannoprotein; plays a role in maintenance of newly synthesized areas of cell wall; localizes to periphery of small buds, septum region of larger buds, and shmoo tip; CCW12 has a paralog, YDR134C, that arose from the whole genome duplication [Source:SGD;Acc:S000004100] 3.5581097 9.2203435 0.3858977
## 4 1028 -3.252 -5.057292 0.4411939 37.13126 3.360154e-11 0.033977287 YKL081W 0.2490052 104942 TEF4 protein_coding Gamma subunit of translational elongation factor eEF1B; stimulates the binding of aminoacyl-tRNA (AA-tRNA) to ribosomes by releasing eEF1A (Tef1p/Tef2p) from the ribosomal complex [Source:SGD;Acc:S000001564] 10.5467254 3.3365413 3.1609755
## 5 171 -3.148 -5.410312 0.5630907 37.13126 3.910600e-09 0.044535051 YBR135W 0.2460829 3619 CKS1 protein_coding Cyclin-dependent protein kinase regulatory subunit and adaptor; interacts with Cdc28p(Cdk1p); required for G1/S and G2/M phase transitions and budding; mediates the phosphorylation and degradation of Sic1p; modulates proteolysis of M-phase targets through interactions with the proteasome; role in transcriptional regulation, recruiting proteasomal subunits to target gene promoters [Source:SGD;Acc:S000000339] 0.3501230 0.5847066 0.5988012
## 6 216 2.230 5.580881 0.8532788 37.13126 7.417209e-06 -0.033413526 YBR269C 0.2496553 2289 SDH8 protein_coding Protein required for assembly of succinate dehydrogenase; interacts with flavinylated Sdh1p and may function as a chaperone for free Sdh1p, protecting its FAD cofactor from redox reactions before assembly of the complex; soluble protein of the mitochondrial matrix; respiratory defect of null mutant is functionally complemented by Drosophila and human orthologs [Source:SGD;Acc:S000000473] 0.8487821 0.5038261 1.6846727
## 7 451 -2.230 -4.340305 0.5408115 37.13126 1.874030e-07 0.055220190 YDR363W-A 0.1454215 21193 SEM1 protein_coding Component of lid subcomplex of 26S proteasome regulatory subunit; involved in mRNA export mediated by TREX-2 complex (Sac3p-Thp1p); assumes different conformations in different contexts, functions as molecular glue stabilizing the Rpn3p/Rpn7p regulatory heterodimer, and tethers it to lid helical bundle; ortholog of human DSS1; protein abundance increases in response to DNA replication stress [Source:SGD;Acc:S000007235] 3.0505759 0.6673323 4.5712993
## 8 1410 -2.212 -3.739133 0.3960689 37.13126 4.685548e-09 0.015982241 YNL067W 0.2809500 30030 RPL9B protein_coding Ribosomal 60S subunit protein L9B; homologous to mammalian ribosomal protein L9 and bacterial L6; RPL9B has a paralog, RPL9A, that arose from a single-locus duplication [Source:SGD;Acc:S000005011] 1.7036378 1.8831046 0.9046963
## 9 1422 2.161 4.458987 0.6021330 37.13126 8.077497e-07 0.004611877 YNL098C 0.2404676 7185 RAS2 protein_coding GTP-binding protein; regulates nitrogen starvation response, sporulation, and filamentous growth; farnesylation and palmitoylation required for activity and localization to plasma membrane; homolog of mammalian Ras proto-oncogenes; RAS2 has a paralog, RAS1, that arose from the whole genome duplication [Source:SGD;Acc:S000005042] 1.2837028 0.7559926 1.6980362
## 10 1125 2.111 3.538636 0.3775617 37.13126 5.045681e-09 -0.001872867 YLR027C 0.2498340 12186 AAT2 protein_coding Cytosolic aspartate aminotransferase involved in nitrogen metabolism; localizes to peroxisomes in oleate-grown cells [Source:SGD;Acc:S000004017] 0.6183686 1.0157893 0.6087568
## ...
## 160 of 1805 non-zero log2 fold change at FDR 0.05
## Prior df 21.1Gene loadings for C3
limma_confects(fit_comp, "C3", full=TRUE, fdr=0.05)## $table
## rank index confect effect se df fdr_zero AveExpr name per_read_var total_reads symbol biotype product dispersion_before dispersion_trend dispersion_after
## 1 614 -1.628 -4.269298 0.5878671 37.13126 7.491684e-06 0.015324847 YFR052C-A 0.14797847 4967 <NA> protein_coding Dubious open reading frame; unlikely to encode a functional protein, based on available experimental and comparative sequence data; overlaps ORF HXK1/YFR053C [Source:SGD;Acc:S000028768] 2.0850412 0.3442802 6.0562329
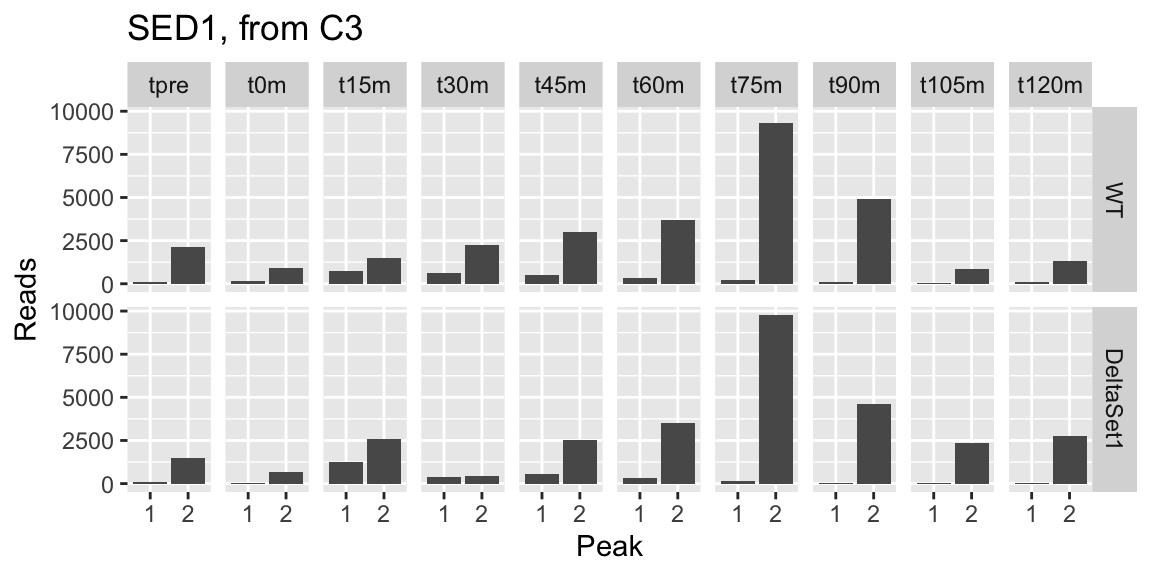
## 2 355 1.628 3.056850 0.3304292 37.13126 6.392750e-08 -0.027501019 YDR077W 0.07946431 66406 SED1 protein_coding Major stress-induced structural GPI-cell wall glycoprotein; associates with translating ribosomes, possible role in mitochondrial genome maintenance; ORF contains two distinct variable minisatellites; SED1 has a paralog, SPI1, that arose from the whole genome duplication [Source:SGD;Acc:S000002484] 8.9434859 0.5784954 15.4599072
## 3 175 1.369 2.661694 0.3086568 37.13126 1.935240e-07 0.025164735 YBR149W 0.19813807 9498 ARA1 protein_coding NADP+ dependent arabinose dehydrogenase; involved in carbohydrate metabolism; purified as homodimer; naturally occurs with a N-terminus degradation product [Source:SGD;Acc:S000000353] 0.4811818 0.6685896 0.7196968
## 4 20 1.320 4.356591 0.7420397 37.13126 2.091532e-04 0.029427305 snR40 0.26487599 3100 <NA> snoRNA C/D box small nucleolar RNA (snoRNA); guides 2'-O-methylation of large subunit (LSU) rRNA at position U898 and small subunit (SSU) rRNA at position G1271 [Source:SGD;Acc:S000007304] 1.0915309 0.6060608 1.8010255
## 5 1337 1.267 3.783609 0.6265230 37.13126 1.650253e-04 0.011229528 YMR194C-B 0.23747967 2270 CMC4 protein_coding Protein that localizes to the mitochondrial intermembrane space; localizes via the Mia40p-Erv1p system; contains twin cysteine-x(9)-cysteine motifs [Source:SGD;Acc:S000028514] 0.6970526 0.4709791 1.4800075
## 6 1422 -1.123 -3.024115 0.4807299 37.13126 9.033870e-05 0.004611877 YNL098C 0.24046761 7185 RAS2 protein_coding GTP-binding protein; regulates nitrogen starvation response, sporulation, and filamentous growth; farnesylation and palmitoylation required for activity and localization to plasma membrane; homolog of mammalian Ras proto-oncogenes; RAS2 has a paralog, RAS1, that arose from the whole genome duplication [Source:SGD;Acc:S000005042] 1.2837028 0.7559926 1.6980362
## 7 1395 0.998 3.076415 0.5326826 37.13126 2.507792e-04 0.063641680 YNL036W 0.24993851 5866 NCE103 protein_coding Carbonic anhydrase; metalloenzyme that catalyzes CO2 hydration to bicarbonate, which is an important metabolic substrate, and protons; not expressed under conditions of high CO2, such as inside a growing colony, but transcription is induced in response to low CO2 levels, such as on the colony surface in ambient air; poorly transcribed under aerobic conditions and at an undetectable level under anaerobic conditions; abundance increases in response to DNA replication stress [Source:SGD;Acc:S000004981] 1.8347943 0.7271858 2.5231437
## 8 1052 -0.934 -3.691046 0.7149819 37.13126 1.269727e-03 -0.043991761 YKL163W 0.13916375 808 PIR3 protein_coding O-glycosylated covalently-bound cell wall protein; required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway; coding sequence contains length polymorphisms in different strains; PIR3 has a paralog, HSP150, that arose from the whole genome duplication [Source:SGD;Acc:S000001646] 0.3997888 0.1842873 2.1693776
## 9 56 0.791 7.112280 1.6442357 36.13126 6.471066e-03 0.174639982 tP(UGG)N1 0.23279608 507 <NA> tRNA Proline tRNA (tRNA-Pro), predicted by tRNAscan-SE analysis; target of K. lactis zymocin [Source:SGD;Acc:S000006685] 0.9548050 0.3415671 2.7953660
## 10 769 0.772 3.665236 0.7654125 37.13126 2.852370e-03 -0.023851955 YGR243W 0.23448147 1172 MPC3 protein_coding Highly conserved subunit of mitochondrial pyruvate carrier; more highly expressed in glucose-containing minimal medium than in lactate-containing medium; expression regulated by osmotic and alkaline stresses; protein abundance increases in response to DNA replication stress; MPC3 has a paralog, MPC2, that arose from the whole genome duplication [Source:SGD;Acc:S000003475] 0.4380471 0.3856953 1.1357336
## ...
## 113 of 1805 non-zero log2 fold change at FDR 0.05
## Prior df 21.1Gene loadings for C4
limma_confects(fit_comp, "C4", full=TRUE, fdr=0.05)## $table
## rank index confect effect se df fdr_zero AveExpr name per_read_var total_reads symbol biotype product dispersion_before dispersion_trend dispersion_after
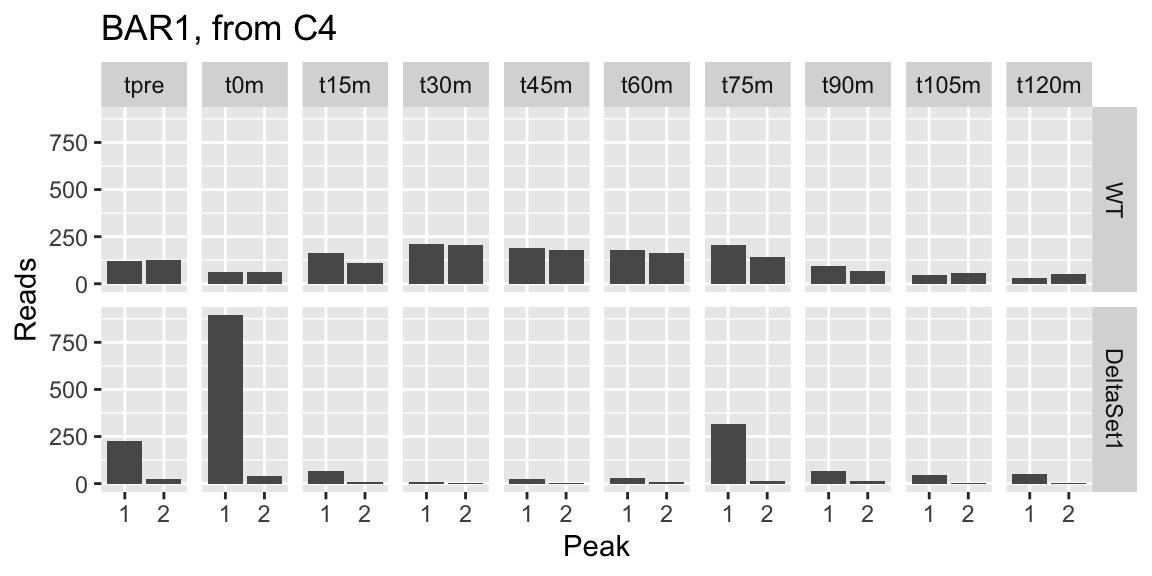
## 1 860 4.360 7.184817 0.6203456 37.13126 6.208646e-11 0.008910808 YIL015W 0.2089827 4313 BAR1 protein_coding Aspartyl protease; secreted into the periplasmic space of mating type a cell; helps cells find mating partners; cleaves and inactivates alpha factor allowing cells to recover from alpha-factor-induced cell cycle arrest [Source:SGD;Acc:S000001277] 1.5452035 0.5075547 3.0444081
## 2 6 -4.140 -6.499207 0.5455377 37.13126 5.408596e-11 0.050189640 RDN5-1 0.2394686 4029 <NA> rRNA 5S ribosomal RNA (5S rRNA); only complete sequence of 6 repeated RDN5 alleles; able to support viability when provided as sole form of 5S rRNA; component of the large (60S) ribosomal subunit; localized to the nucleolus via interaction with Rpl5p; may play a role in translational frame fidelity; transcription is mediated by PolIII and activated by TFIIIA and TFIIIE [Source:SGD;Acc:S000006479] 1.1924250 0.5888775 2.0249117
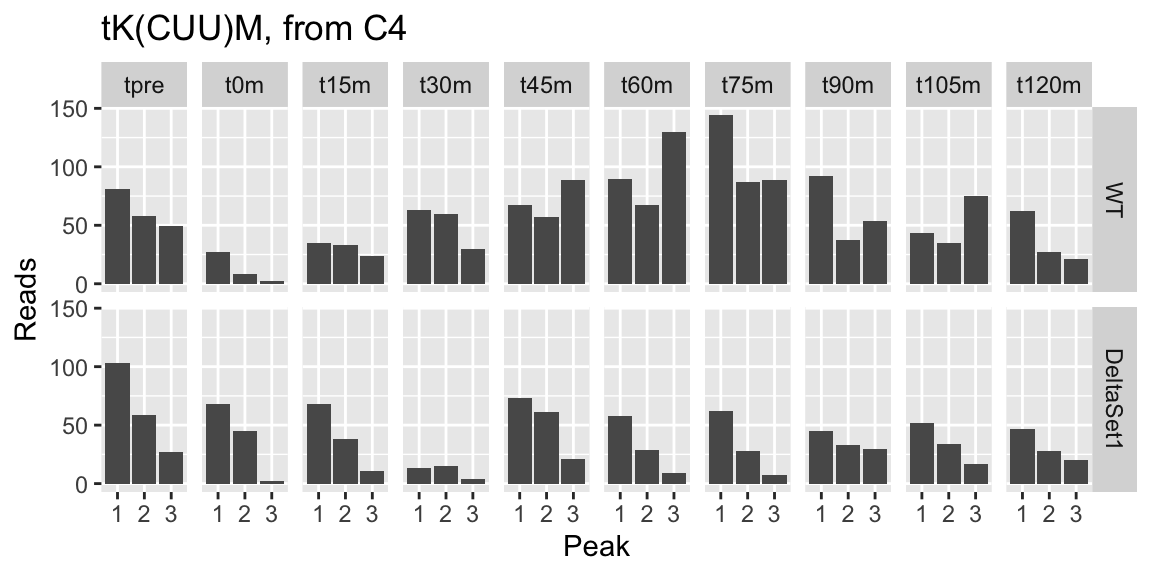
## 3 49 1.492 4.561583 0.7329322 37.13126 9.274286e-05 -0.041137359 tK(CUU)M 0.2881947 2843 <NA> tRNA Lysine tRNA (tRNA-Lys), predicted by tRNAscan-SE analysis; a small portion is imported into mitochondria via interaction with mt lysyl-tRNA synthetase Msk1p and is necessary to decode AAG codons at high temperature, when base modification of mt-encoded tRNA-Lys is reduced [Source:SGD;Acc:S000006627] 1.3642565 0.6545660 2.0842154
## 4 1201 1.429 4.429143 0.7332200 37.13126 9.508451e-05 0.014684787 YLR286C 0.2423945 1548 CTS1 protein_coding Endochitinase; required for cell separation after mitosis; transcriptional activation during the G1 phase of the cell cycle is mediated by transcription factor Ace2p [Source:SGD;Acc:S000004276] 0.6292428 0.4304440 1.4618459
## 5 47 -1.242 -3.249218 0.4996632 37.13126 4.669415e-05 -0.048926352 tK(CUU)J 0.0900000 1200 <NA> tRNA Lysine tRNA (tRNA-Lys), predicted by tRNAscan-SE analysis; a small portion is imported into mitochondria via interaction with mt lysyl-tRNA synthetase Msk1p and is necessary to decode AAG codons at high temperature, when base modification of mt-encoded tRNA-Lys is reduced [Source:SGD;Acc:S000006625] 0.3220354 0.1124589 2.8635829
## 6 1125 -1.129 -2.413699 0.3248468 37.13126 3.369473e-06 -0.001872867 YLR027C 0.2498340 12186 AAT2 protein_coding Cytosolic aspartate aminotransferase involved in nitrogen metabolism; localizes to peroxisomes in oleate-grown cells [Source:SGD;Acc:S000004017] 0.6183686 1.0157893 0.6087568
## 7 58 -1.127 -4.655309 0.9041275 37.13126 5.894600e-04 0.039423397 tP(UGG)O3 0.2500000 892 <NA> tRNA Proline tRNA (tRNA-Pro), predicted by tRNAscan-SE analysis; target of K. lactis zymocin [Source:SGD;Acc:S000006689] 0.5037467 0.3986546 1.2636170
## 8 1422 -1.064 -2.858270 0.4677233 37.13126 9.508451e-05 0.004611877 YNL098C 0.2404676 7185 RAS2 protein_coding GTP-binding protein; regulates nitrogen starvation response, sporulation, and filamentous growth; farnesylation and palmitoylation required for activity and localization to plasma membrane; homolog of mammalian Ras proto-oncogenes; RAS2 has a paralog, RAS1, that arose from the whole genome duplication [Source:SGD;Acc:S000005042] 1.2837028 0.7559926 1.6980362
## 9 921 -1.064 -2.864462 0.4718475 37.13126 9.508451e-05 0.010685254 YJL054W 0.1756457 1918 TIM54 protein_coding Component of the mitochondrial TIM22 complex; involved in insertion of polytopic proteins into the inner membrane [Source:SGD;Acc:S000003590] 0.2100440 0.3020734 0.6953409
## 10 45 -0.992 -4.611214 0.9575052 37.13126 1.205028e-03 0.044953075 tH(GUG)G2 0.2459710 898 <NA> tRNA Histidine tRNA (tRNA-His), predicted by tRNAscan-SE analysis [Source:SGD;Acc:S000006596] 0.5321993 0.3907813 1.3618853
## ...
## 180 of 1805 non-zero log2 fold change at FDR 0.05
## Prior df 21.1Examine individual genes
Let’s examine peak-level read counts for some genes we’ve identified.
examine <- function(gene_wanted, title) {
peak_names <- filter(peaks, gene_name==gene_wanted)$name
counts[peak_names,] %>% melt(value.name="reads", varnames=c("peak","sample")) %>%
left_join(samples, by="sample") %>%
ggplot(aes(x=factor(as.integer(peak)), y=reads)) +
facet_grid(strain ~ time) + geom_col() +
labs(x="Peak",y="Reads",title=title)
}
examine("YLR058C", "SHM2, from C1")